RNA Theraputics
Lipid Nano Particles
RNA therapeutics has rapidly developed since a great success of RNA vaccines against SARS-CoV-2 virus. Encapsulation into Lipid NanoParticles (LNPs) has been proven to be one of the most efficient ways for RNA delivery in vivo. The approach uses lipid mixtures with positively charged cationic lipids that interact to negatively charged RNA molecules, protecting them from from enzymatic degradation and providing intracellular penetration. In a joined effort with Abisch-Frenkel RNA Therapeutics Center (Prof. Igor Ulitsky), we established a publically available facility for RNA-LNP production. Our facility integrates equipment required for LNP assembly, post-assembly processing and quality control. It allows production of minimum 2 ml of RNA-LNPs that covers most of the research needs.
NanoAssembler Ignite:

The Ignite system (Precision NanoSystems) is designed to assemble RNA lipid nano particles particles (LNPs), using microfluidic cartriges. Easy-to-use programming provides high reproducibility of the LNP assembly. Either custom lipids formulations or a commercial kit (GenVoy-ILM) can be applied for RNA encapsulations. Disposable NxGen cartriges eliminates risk of cross-contamination.
Zetasizer:

ZetaSizer Ultra (Malvern Panalytical) is the most advanced analyser enabled to measure (1) size of nanoparticles, (2) particle concentration, and (3) Z-potential of nanoparticles. The device enables to analyse particles up to 1 um. Estimation of particle size and Z-potential is a necessary step of LNP quality control, but can also be used in other applications, like analysis of aggregates.

Typical output of mRNA encapsulation by the GenVoy-ILM lipids. ZetaSizer detects a single pick of 120nms.
Kinetic imaging of cells
Incucyte S3
The Incucyte S3 is an imaging system within an incubator. This enables on-line imaging of cell cultures for time-dependent experiments without disturbing the culture or physical intervention. Images are acquired for up to 6 microtiter plates in all popular SBS-formats. The Incucyte features:
- Fluorescent channel 1 - 480 nm ex /520 nm em
- Fluorescent channel 2 - 585 nm ex /640 nm em
- Phase contrast
- Objectives: 4X/10X/20X
Cell growth assays
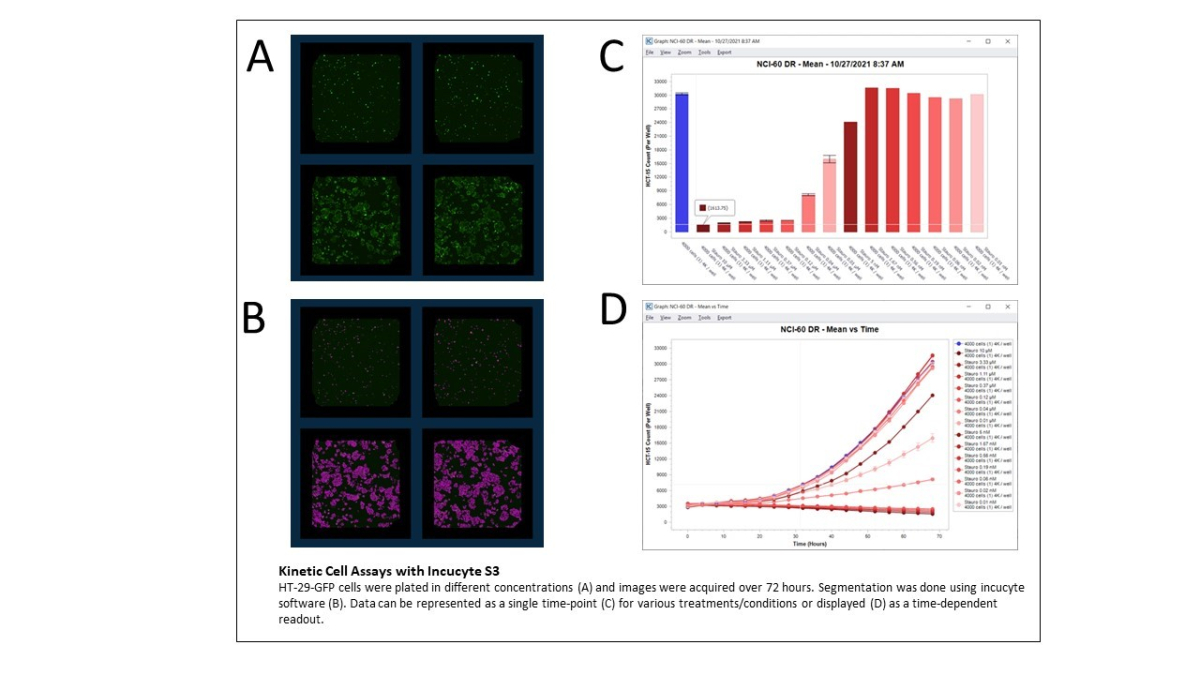
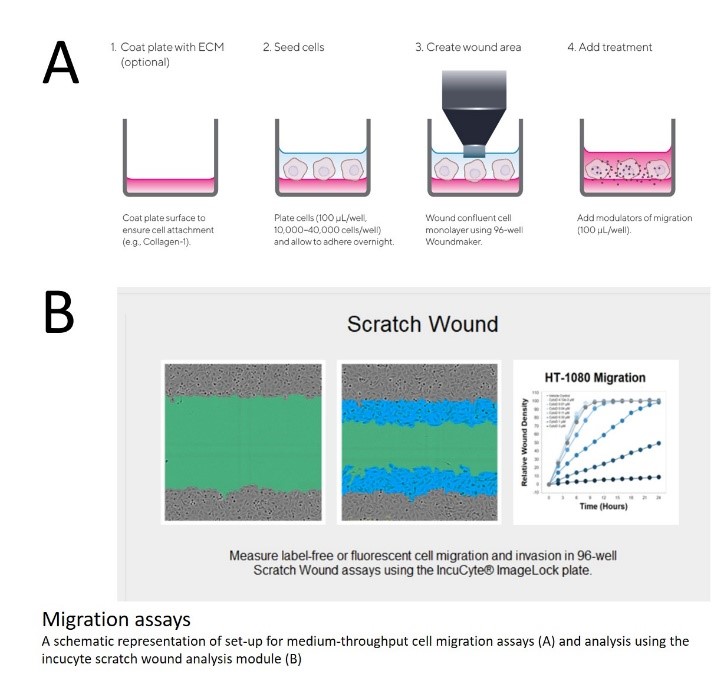
Cell motility assays
Migration of a monolayer is performed on cultures plated in image-lock plates and a modification using the 96-well Woundmaker apparatus to create a calibrated region of interest. Plates and apparatus are available in G-INCPM.
Scratch assay
Contact noga.kozer@weizmann.ac.il
Multiplexed Immunoassays
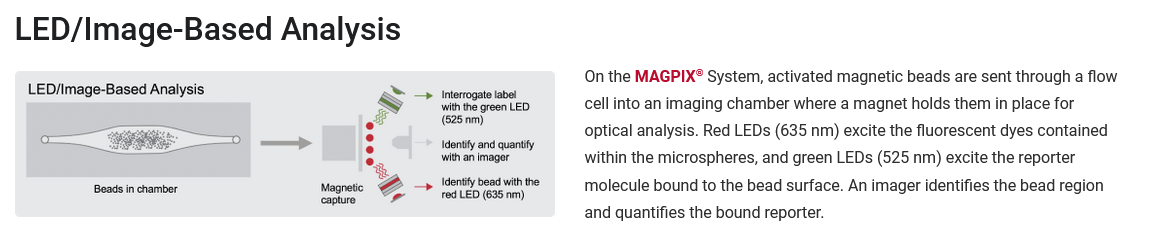
Luminex
Luminex is a bead-based detection platform for multiplexing up to 50 different simultaneous assays in each well. Luminex or alternatively MAPx (Multi-Analyte Profiling, where the “x” represents biomarkers such as proteins, nucleic acids, or polysaccharides that are being tested). xMAP Technology uses labeled microspheres or beads, allowing for the simultaneous capture of multiple analytes from a single reaction. Because of their small size and low density, xMAP microsphere-based assays exhibit virtual solution-phase kinetics during the reaction. The beads are individually read using MAGpix instrument. Additionally, it is possible to automate sample preparation as well. We can perform the whole process or alternatively you can prepare the plates with the relevant beads and samples and then bring it to read in our center. There are several analytes, for example, cytokines or antibodies against pathogens that are already set up in existing commercial kits. If a commercial kit is not available, we can help you in customizing your beads to enable testing of relevant analytes.
Recent publications from our group using this technology
https://www.jci.org/articles/view/150319
Contact leonardo.solmesky@weizmann.ac.il for more information
Target-engagement assays
Compound-induced thermal stabilization of proteins is frequently used to confirm binding of small molecule inhibitors to protein targets.
Thermal Shift Assay
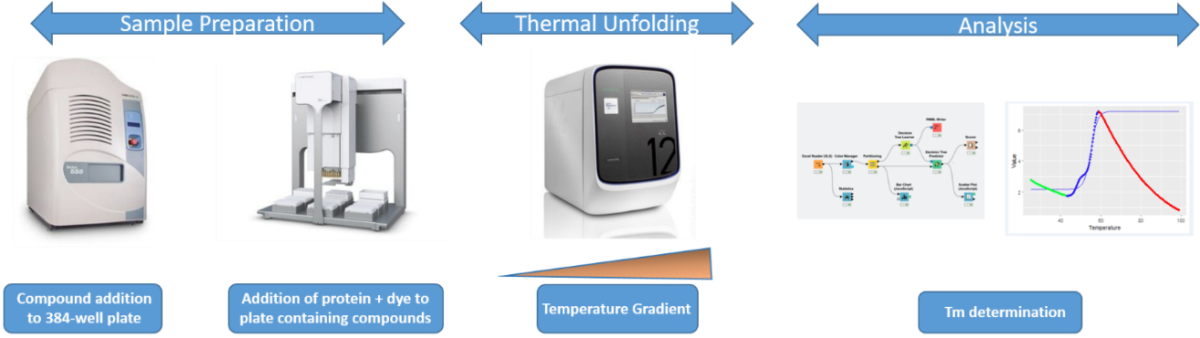
Protein stability screenings can be performed using a protein melting method. These can reveal changes in protein structure as a result of their thermal stability. Thermal shift assays (TSA) have traditionally been applied to purified proteins to study their thermal stabilization by the use of dyes with sensitive fluorescence detection (for e.g. SYPRO Orange, Glo-Melt) that enable rapid and relative inexpensive characterization by using a real-time PCR instrument. By screening a wide range of solutions and additives in a 384-well format, we can easily identify conditions that perturb the stability of recombinant proteins. The workflow below (Figure 1) shows our unit’s established methodology for a high-throughput thermal denaturation of a recombinant protein by using a fluorescent dye.
Cell-based thermal shift assay (CETSA)
In recent years, the thermal shift assay armamentaria evolved to include a cellular thermal shift assay (CETSA). Though based on the same principle as conventional TSAs, CETSA allows for ligand-induced stabilization in more complex environments like intact cells or cell lysates. This technology is currently offered in our unit for target engagement confirmation following a high throughput screening. This method can currently be applied to small numbers of compounds of high interest and potency.
Recent publications from our group using this technology
https://www.nature.com/articles/s41598-020-77028-8
Contact silvia.carvalho@weizmann.ac.il
Fluorescent-labelled cell panel
Profiling bioactivity of a compound across multiple cell lines it is useful to compare effects in diverse genetic context. We collaborated with the Stem Cell Unit of LSCF to generate an easy-to-use resource of 20 cell lines from different tissue origins, each expressing a stable fluorescent marker. All cell lines have been calibrated with staurosporine as a reference compound. GFP was introduced to cell lines using TALENS according to https://doi.org/10.1002/stem.1653 selected by FACS and propagated in culture.
Recent publications from our group using this technology
https://pubs.acs.org/doi/abs/10.1021/jacs.2c00269
Contact noga.kozer@weizmann.ac.il